Readings: "Alternative trans-splicing: a novel mode of pre-mRNA processing" by Horiuchi et al
Reading Horiuchi et al: Alternative trans-splicing: a novel mode of pre-mRNA processing
Biology of the Cell, Volume 98, Issue 2, 2006
http://www.ncbi.nlm.nih.gov/pubmed/16417469
Horiuchi's paper summarizes a number of findings concerning the subject of trans-splicing.
Trans-splicing is a mechanism that naturally occurs in cells. It has also been experimentally used to correct genetic defects in mice, so it is both an interesting natural phenomenon and a possible avenue of investigation for therapeutic applications.
Splicing, inside a cell, is one of the numerous steps from DNA to RNA to protein. Splicing operates on premature mRNA (pre-mRNA for short), from which it excises introns and joins the remaining exons back together. Splicing comes in two varieties: cis and trans. Cis-splicing joins exons within a single pre-mRNA molecule, while trans-splicing joins exons from different pre-mRNA molecules. Trans-splicing seems to be rare compared to cis-splicing, but it has been documented for a number of organisms, including unicellulars, the roundworm C. elegans, Drosophila, rat, and human. In unicellulars, all mRNAs are prepended with an SL (spliced leader) RNA through trans-splicing. This process of SL addition is well documented. Note that it does not expand the diversity of the proteome of the organism, since the SL sequence is non-coding.
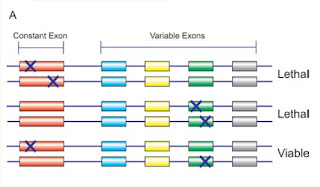
Things may be different in the higher organisms in which trans-splicing has been documented, where the identified cases do not follow the pattern of SL addition. A functional signficance of trans-splicing has only been established for some of these cases - for others the question of functional significance remains open, as does the question of how frequent trans-splicing events are in different species or how exactly trans-splicing works. In the case of the lola gene in Drosophila, trans-splicing was studied by observing the effects of targeted interallelic mutation. A schematic of the experimens and their results is shown in the following figure:
Depicted are three experiments, each with one introduced mutation per allele. The viability of the bottom-most experiment can be explained by interallelic trans-splicing of the intact exons.
The authors also speculate on a possible conditional advantage of trans-splicing over cis-splicing:
In the cis-splicing mode, the proper expression of isoforms requires strict regulation of all potential polyadenylation sites in coordination with the alternative splicing. The regulatory mechanism would be enormously complex if the gene contains as many exons as mod(mdg4) or lola. Trans-splicing may overcome this problem, allowing for the joining of exons between two discontinuous transcripts without any mechanism that would select a particular polyadenylation site to produce a given isoform.So far, no particular genomic "signal" sequences are known that would let us recognize trans-splicing sites from the genomic sequence alone - for all we know at the moment, the splice sites of trans-spliced exons look just like those of cis-spliced ones.
One hypothesis on the mechanism of trans-splicing hinges on the observation that in vitro trans-splicing between synthetic pre-mRNAs is enhanced when intronic regions of the pre-mRNA molecules contain complementary sequences. Watson-Crick base-pairing between the pre-mRNAs could indeed explain how trans-splicing is enabled or at least facilitated.

Comments
Post a Comment